Anti-SARS-CoV-2 Spike RBD (Clone 2165) – HRP
Anti-SARS-CoV-2 Spike RBD (Clone 2165) – HRP
Product No.: LT1910
- -
- -
Product No.LT1910 Clone 2165 Target SARS-CoV-2 RBD Product Type Recombinant Monoclonal Antibody Alternate Names COV2-2165, SARS-CoV-2 Spike RBD Antibody, Receptor Binding Domain Monoclonal Antibody Isotype Human IgG1 Applications ELISA , IHC |
Data
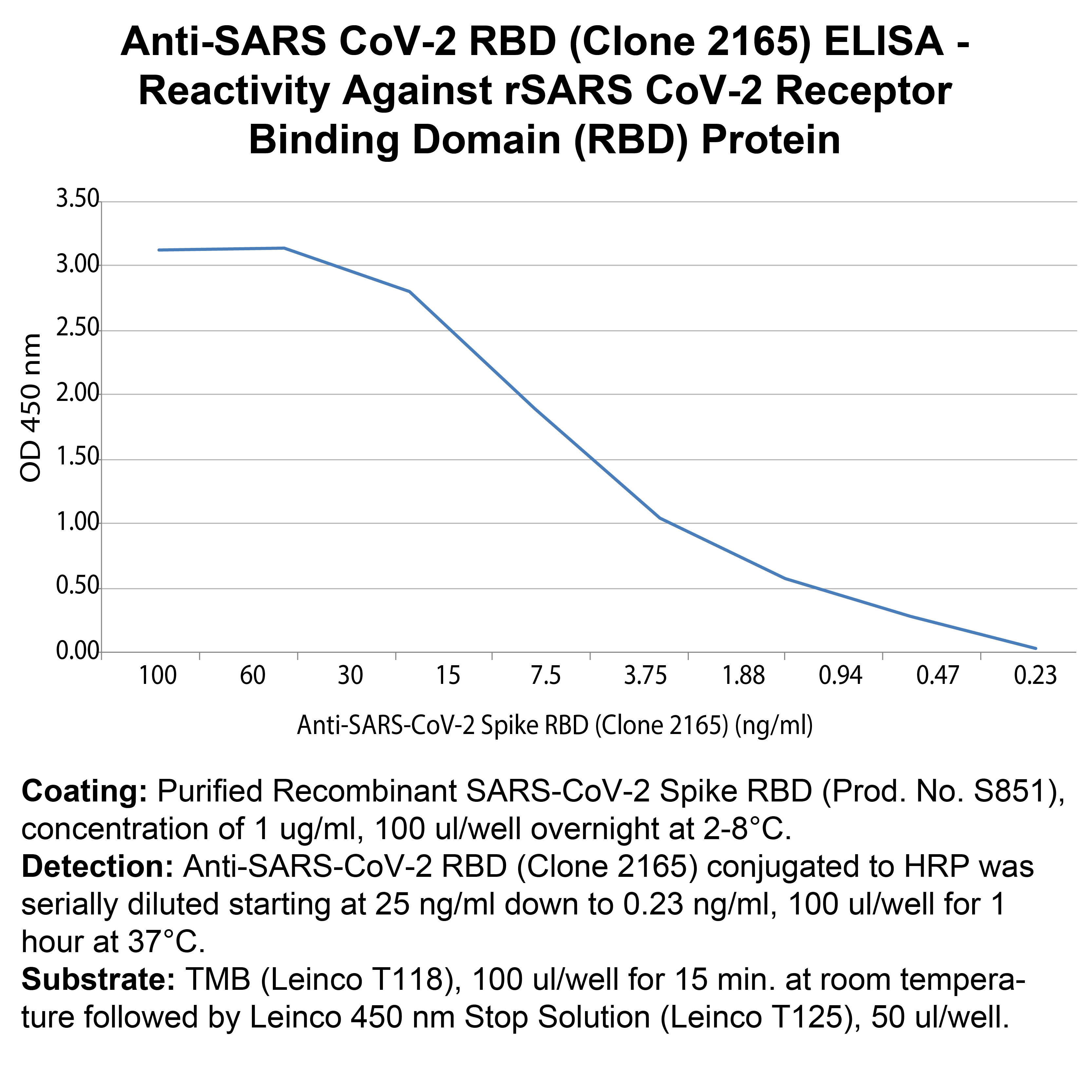 Coating: Purified Recombinant SARS-CoV-2 Spike RBD (Prod. No. S851), concentration of 1 ug/ml, 100 ul/well overnight at 2-8°C.
Detection: Anti-SARS-CoV-2 RBD (Clone 2165) conjugated to HRP was serially diluted starting at 25 ng/ml down to 0.23 ng/ml, 100 ul/well for 1 hour at 37°C.
Substrate: TMB (Leinco T118), 100 ul/well for 15 min. at room temperature followed by Leinco 450 nm Stop Solution (Leinco T125), 50 ul/well.
Coating: Purified Recombinant SARS-CoV-2 Spike RBD (Prod. No. S851), concentration of 1 ug/ml, 100 ul/well overnight at 2-8°C.
Detection: Anti-SARS-CoV-2 RBD (Clone 2165) conjugated to HRP was serially diluted starting at 25 ng/ml down to 0.23 ng/ml, 100 ul/well for 1 hour at 37°C.
Substrate: TMB (Leinco T118), 100 ul/well for 15 min. at room temperature followed by Leinco 450 nm Stop Solution (Leinco T125), 50 ul/well. - -
- -
Antibody DetailsProduct DetailsReactive Species SARS-CoV-2 ⋅ Virus Expression Host HEK-293 Cells Immunogen Sequenced from human survivors of COVID-19 (SARS-CoV-2) Product Concentration 0.5 mg/ml Formulation This recombinant HRP-conjugated antibody is formulated in 0.01 M phosphate buffered saline (150 mM NaCl) PBS pH 7.2 - 7.4, 1% BSA. (Warning: Use of sodium azide as a preservative will inhibit the enzyme activity of horseradish peroxidase) Storage and Handling This horseradish peroxidase conjugated monoclonal antibody is stable when stored at 2-8°C. Do not freeze. Country of Origin USA Shipping Overnight on Blue Ice. RRIDAB_2893934 Applications and Recommended Usage? Quality Tested by Leinco ELISA Additional Applications Reported In Literature ? IHC Each investigator should determine their own optimal working dilution for specific applications. See directions on lot specific datasheets, as information may periodically change. DescriptionDescriptionSpecificity Anti-SARS-CoV-2 Spike RBD-HRP, clone 2165, specifically targets an epitope on the SARS-CoV-2 spike protein receptor-binding domain (RBD). Background Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), is an enveloped, single-stranded, positive-sense RNA virus that belongs to the Coronaviridae family 1. The SARS-CoV-2 genome, which shares 79.6% identity with SARS-CoV, encodes four essential structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid protein (N) 2. The S protein is a transmembrane, homotrimeric, class I fusion glycoprotein that mediates viral attachment, fusion, and entry into host cells 3. Each ~180 kDa monomer contains two functional subunits, S1 (~700 a.a) and S2 (~600 a.a), that mediate viral attachment and membrane fusion, respectively. S1 contains two major domains, the N-terminal (NTD) and C-terminal domains (CTD). The CTD contains the receptor-binding domain (RBD), which binds to the angiotensin-converting enzyme 2 (ACE2) receptor on host cells 3-5. Although both SARS-CoV and SARS-CoV-2 bind the ACE2 receptor, the RBDs only share ~73% amino acid identity, and the SARS-CoV-2 RBD binds with a higher affinity compared to SARS-CoV 3, 6. The RBD is dynamic and undergoes hinge-like conformational changes, referred to as the “down” or “up” conformations, which hide or expose the receptor-binding motifs, respectively 7. Following receptor binding, S1 destabilizes, and TMPRSS2 cleaves S2, which undergoes a pre- to post-fusion conformation transition, allowing for membrane fusion 8, 9. Polyclonal RBD-specific antibodies can block ACE2 binding 10, 11, and anti-RBD neutralizing antibodies are present in the sera of convalescent COVID19 patients 12, identifying the RBD as an attractive candidate for vaccines and therapeutics. In addition, the RBD is poorly conserved, making it a promising antigen for diagnostic tests 13 14. Serologic tests for the RBD are highly sensitive and specific for detecting SARS-CoV-2 antibodies in COVID19 patients 13 15. Furthermore, the levels of anti-RBD antibodies correlated with SARS-CoV-2 neutralizing antibodies, suggesting the RBD could be used to predict an individual's risk of disease 13. Antigen Distribution The spike RBD is expressed on the surface of SARS-CoV-2. PubMed NCBI Gene Bank ID Research Area COVID-19 . Infectious Disease . Seasonal and Respiratory Infections . Viral . IVD Raw Material References & Citations1. Zhou, P., Yang, X., Wang, X. et al. Nature 579, 270–273. 2020. 2. Wu, F., Zhao, S., Yu, B. et al. Nature 579, 265–269. 2020. 3. Wrapp D, Wang N, Corbett KS, et al. bioRxiv. 2020.02.11.944462. 2020. 4. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Cell. 181(2):281-292.e6. 2020. 5. Li W, Zhang C, Sui J, et al. EMBO J. 24(8):1634-1643. 2005. 6. Shang, J., Ye, G., Shi, K. et al. Nature 581, 221–224. 2020. 7. Gui M, Song W, Zhou H, et al. Cell Res. 27(1):119-129. 2017. 8. Walls AC, Tortorici MA, Snijder J, et al. Proc Natl Acad Sci U S A. 114(42):11157-11162. 2017. 9. Hoffmann M, Kleine-Weber H, Schroeder S, et al. Cell. 181(2):271-280.e8. 2020. 10. Huo J, Zhao Y, Ren J, et al. Cell Host Microbe. S1931-3128(20)30351-6. 2020. 11. Tai, W., He, L., Zhang, X. et al. Cell Mol Immunol 17, 613–620. 2020. 12. Cao Y, Su B, Guo X, et al. Cell. 182(1):73-84.e16. 2020. 13. Premkumar L, Segovia-Chumbez B, Jadi R, et al. medRxiv; 2020. 14. Quinlan BD, Mou H, Zhang L, et al. bioRxiv; 2020. 15. Olba NM, Muller MA, Li W, et al. medRxiv; 2020. Technical ProtocolsCertificate of Analysis |
Formats Available
- -
- -